Here are the basic steps I used to process BioSemi EEG data.
1 load raw data
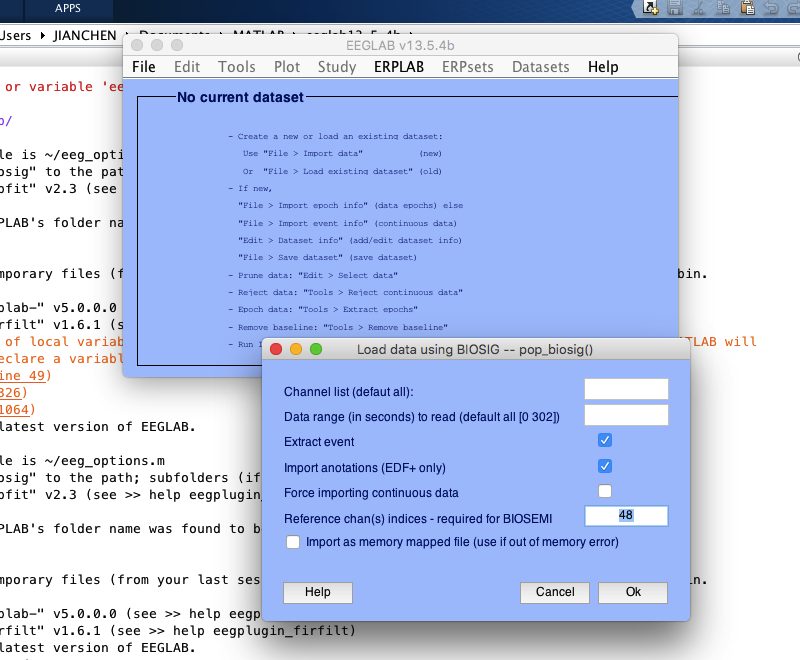
2 channel location
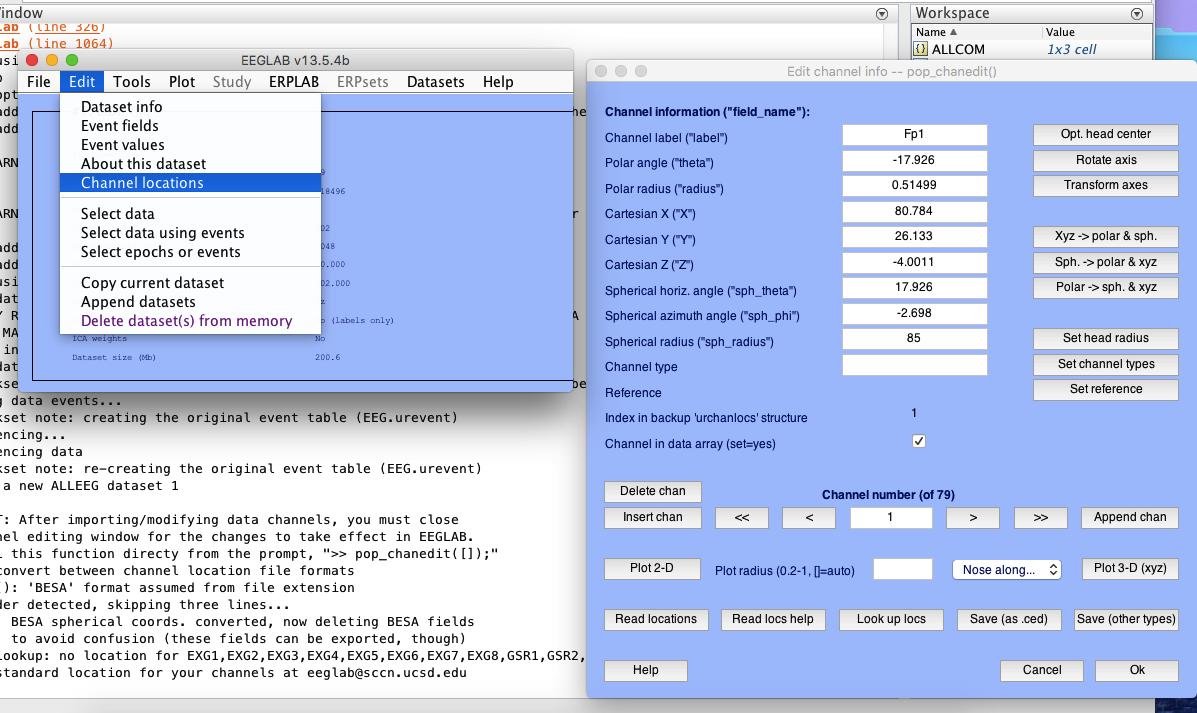
3 re-reference
4 filtering (sometimes the data is very huge and therefore the filtering will be very slow, so we can do the filtering later)
5 Create EEG event list (ERPLAB from now on)
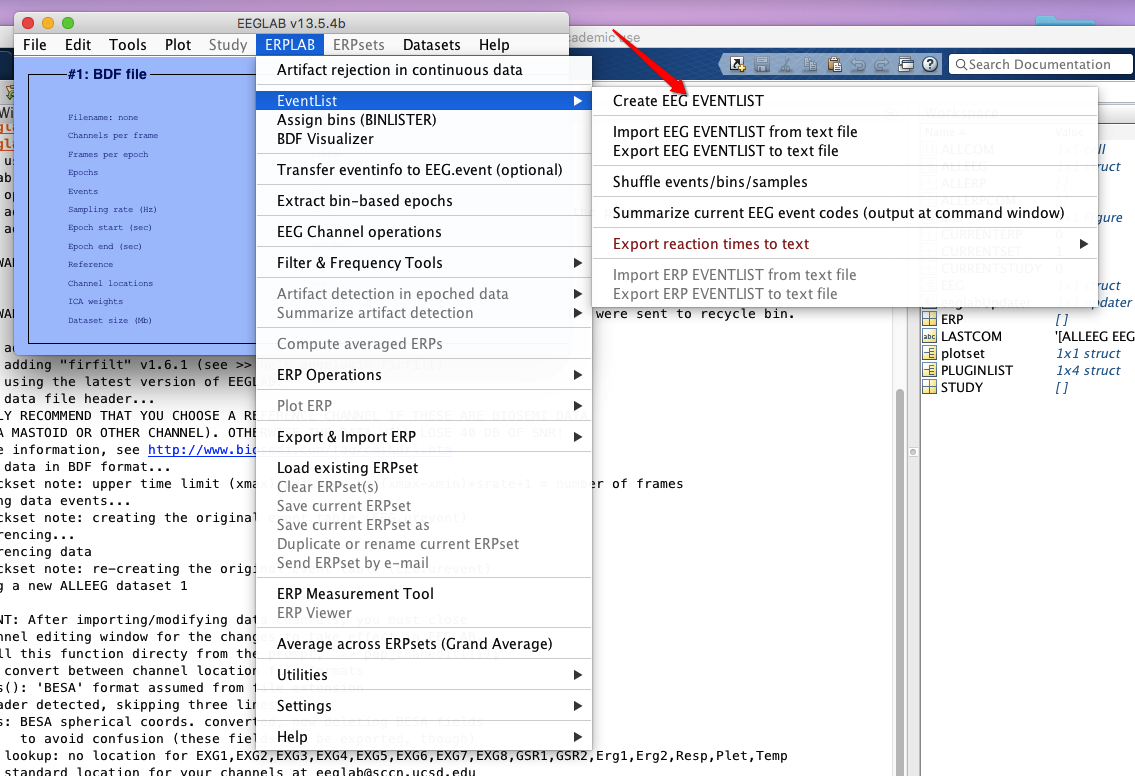
6 Assign bins (how to operate on bins? see here)
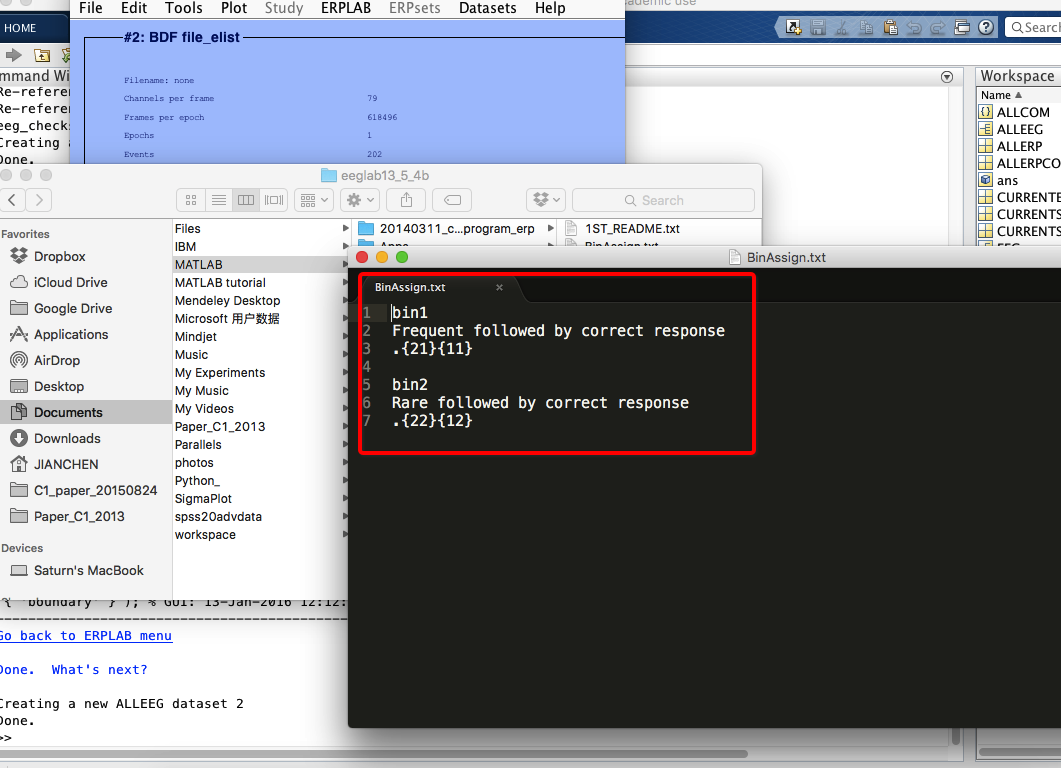
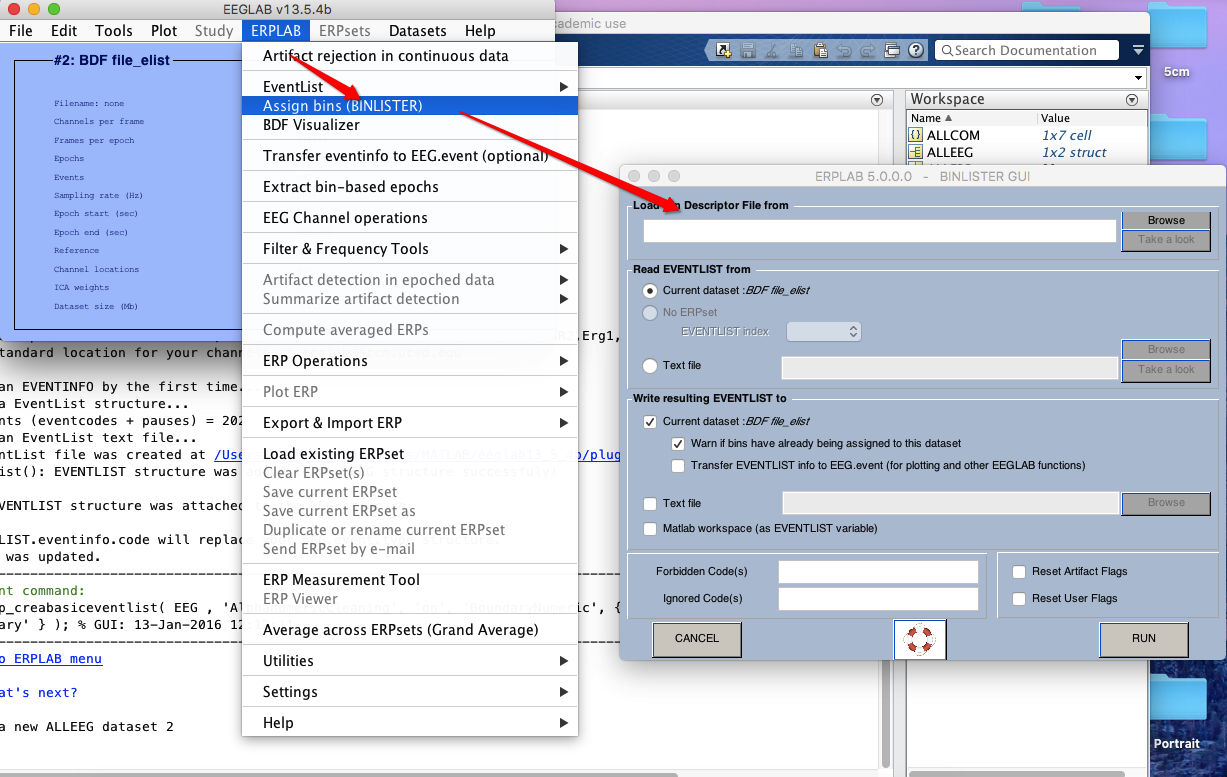
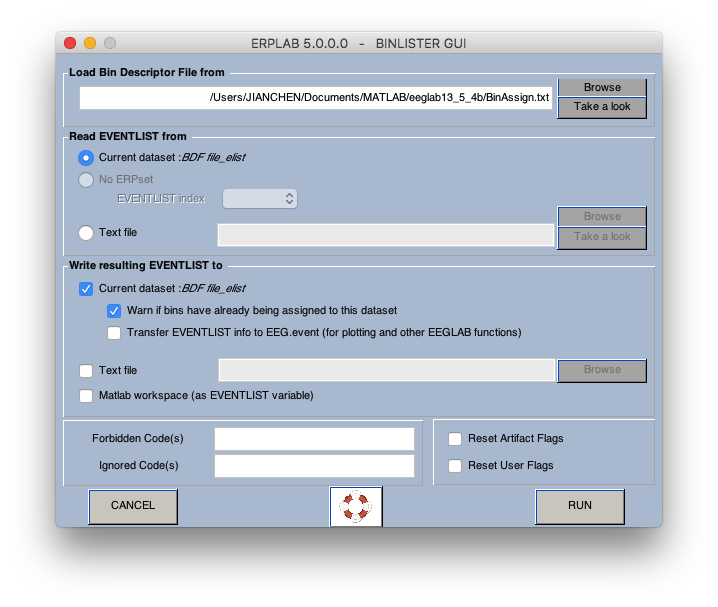
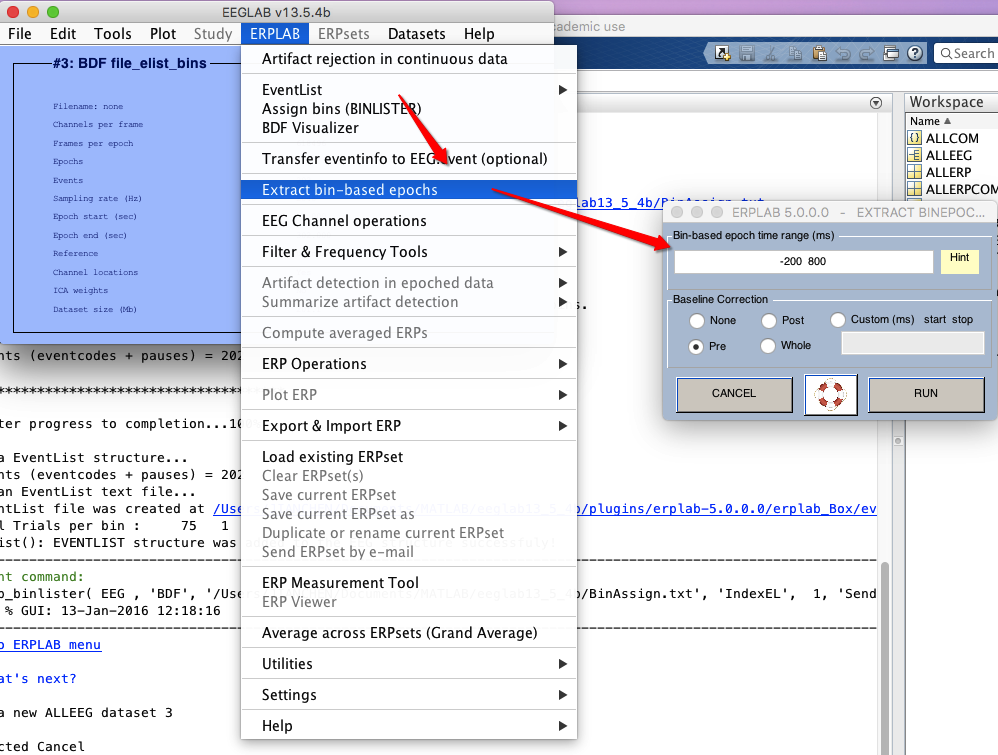
7 extract bin-based epoch
8 artifact rejection
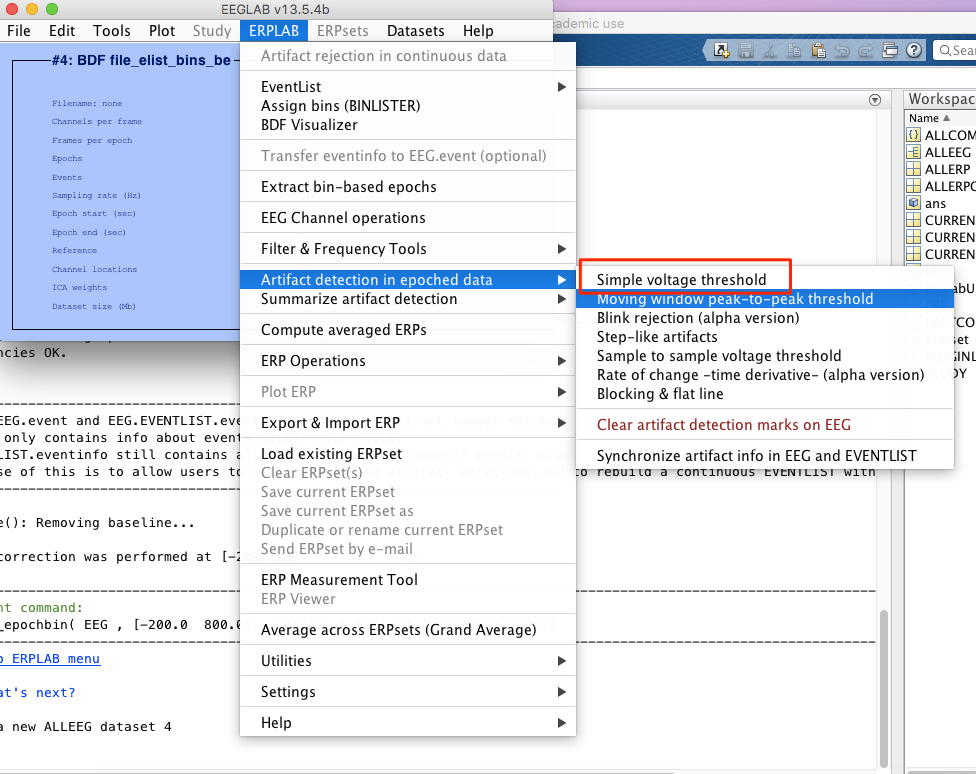
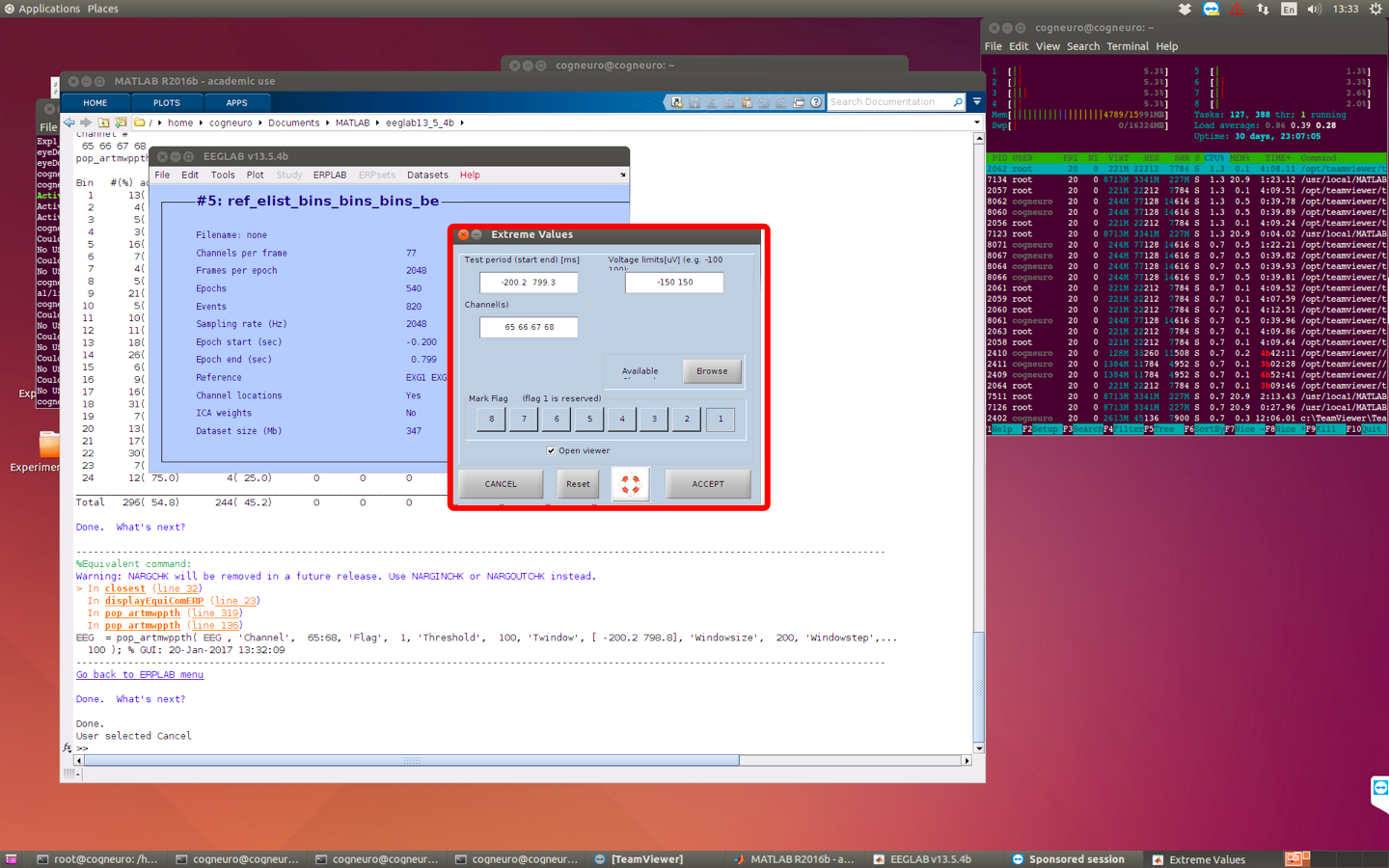
-> choose channel around the eye. -> update and then reject
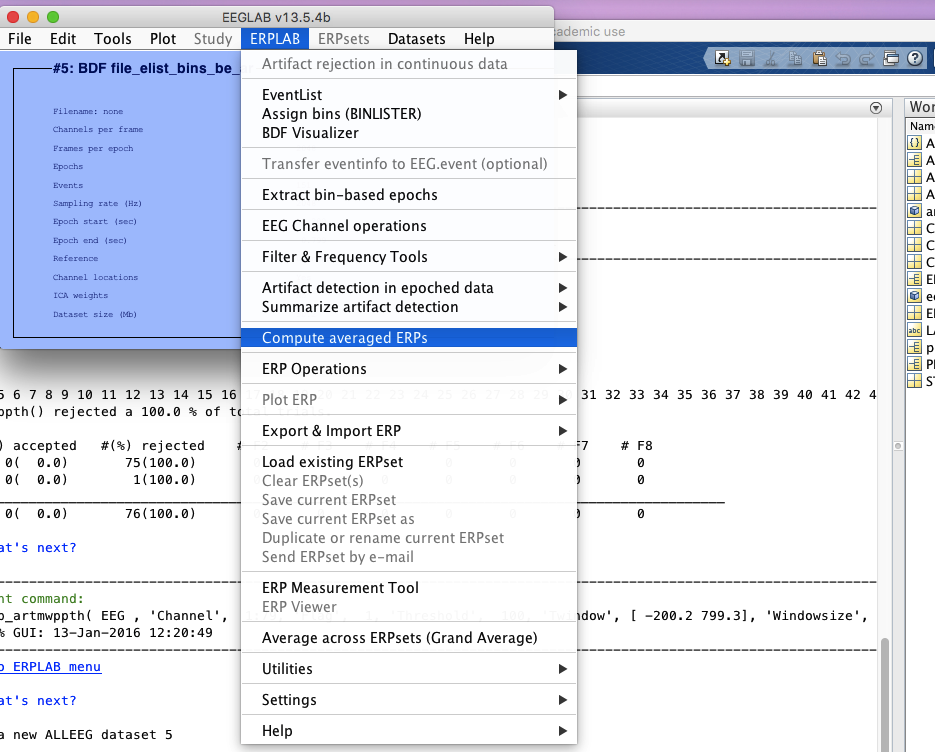
9 compute average erps
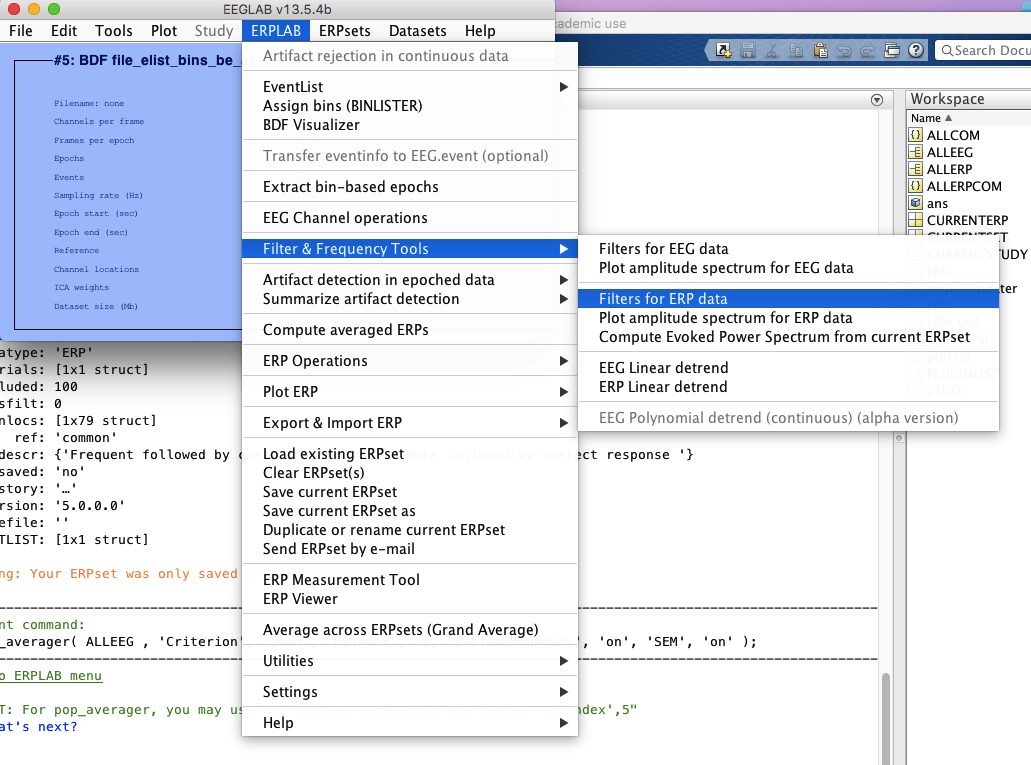
10 filter if you don’t do it in step 4
11 bin operations if necessary
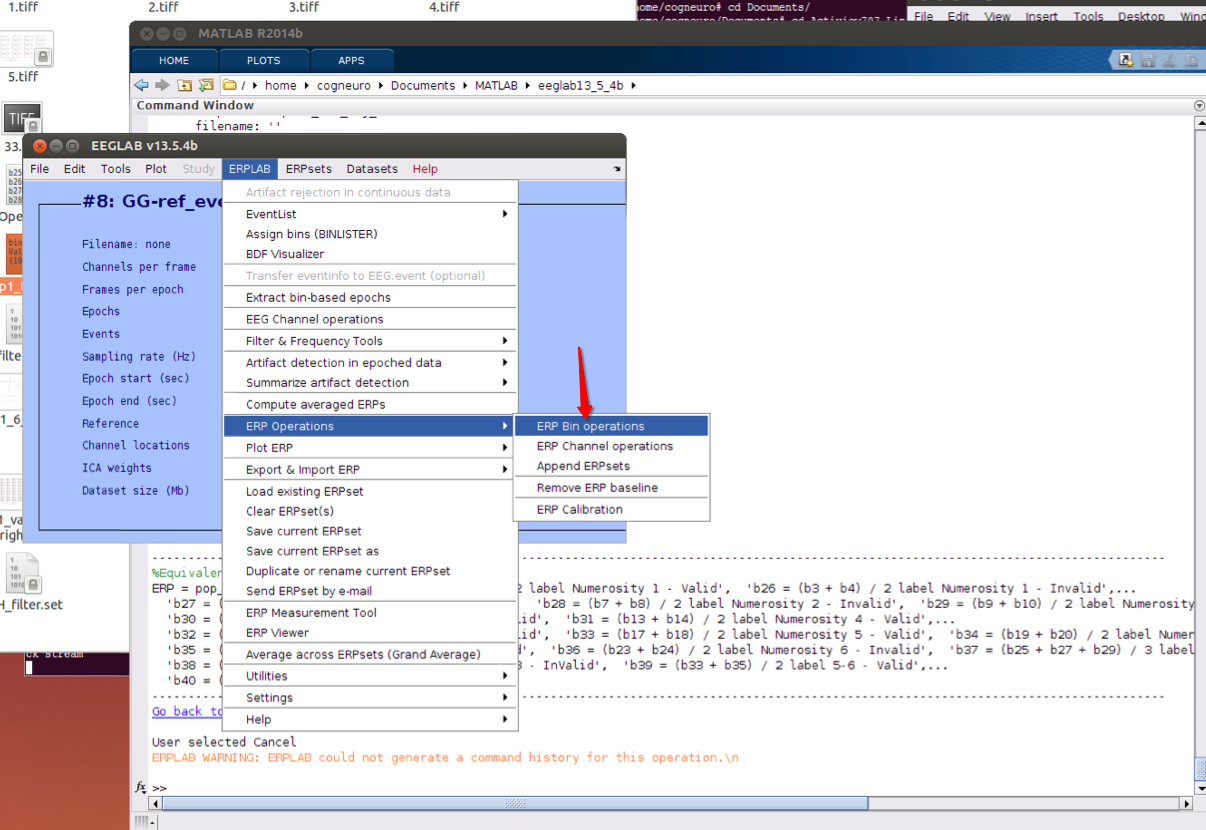
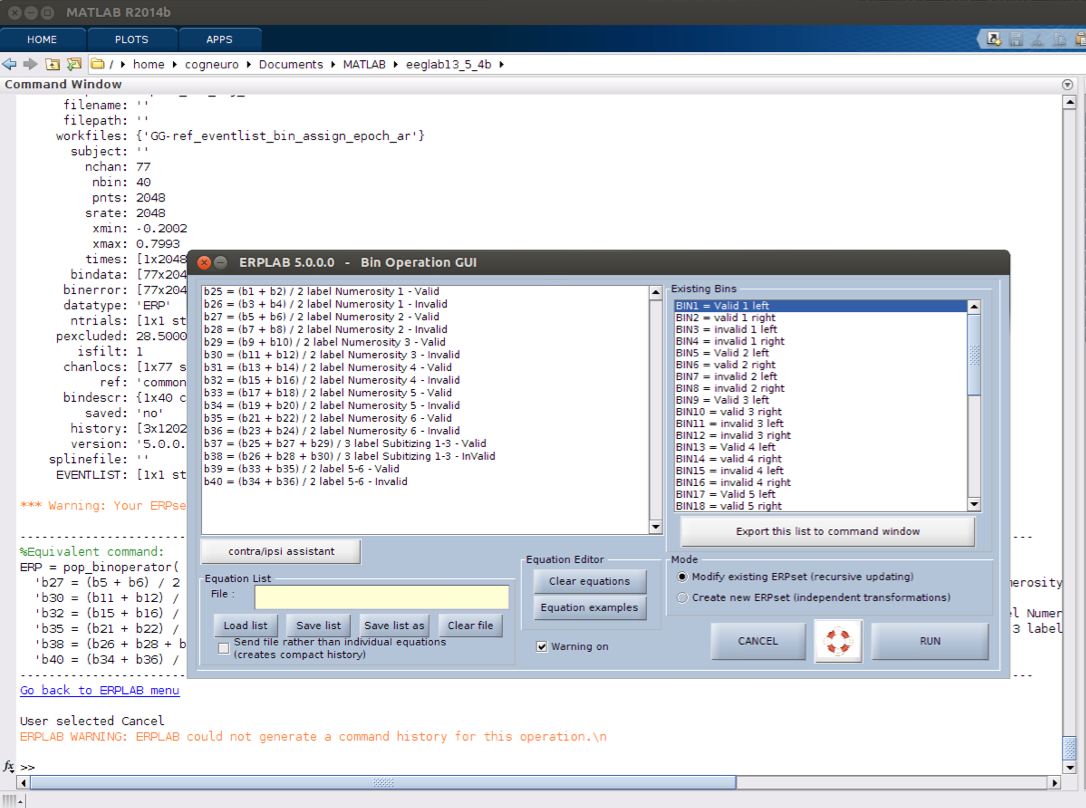
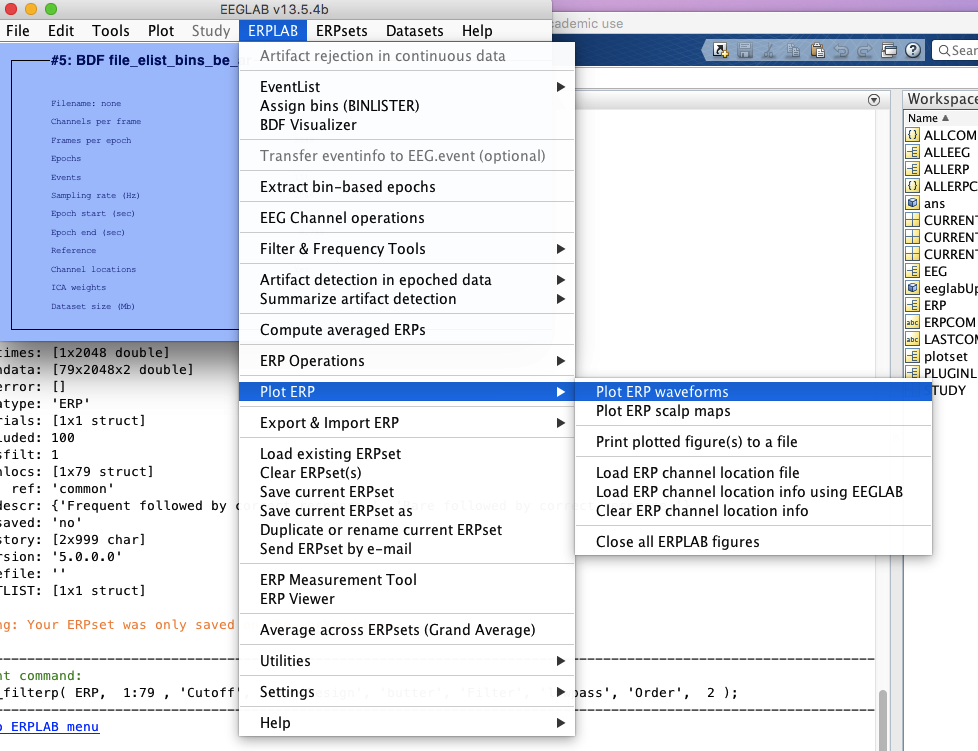
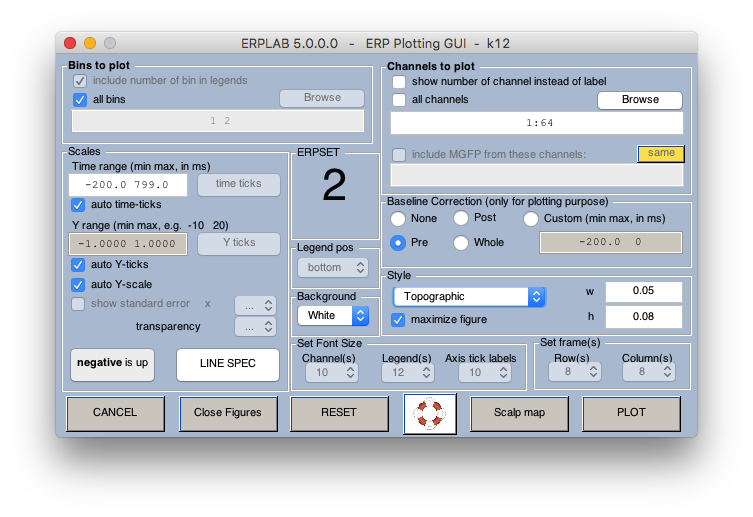
12 Plot ERPs
13 Save figures if you want
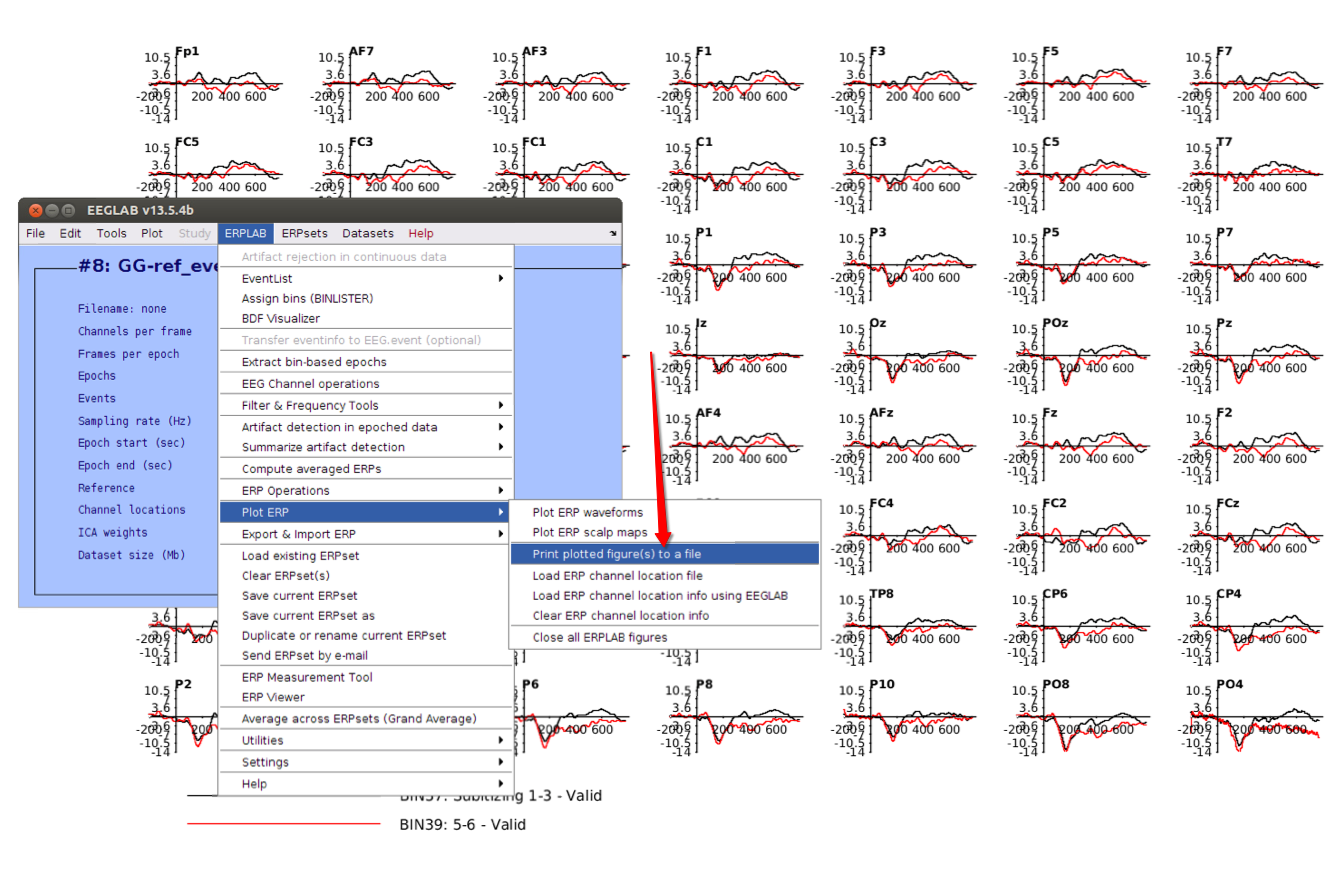
Remember, these are basic steps to illustrate how to process the EEG data using EEGLAB and ERPLAB toolbox, you’re suggested not to strictly follow these steps.